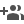The largest genomic study conducted among population groups of Asia has discovered about 200,000 previously unreported novel DNA variants among Asians. The DNA variants are informative points on the DNA or DNA markers. Some of these newly discovered DNA variants have been found in well-studied genes implicated in various diseases, such as diabetes, thalassemia and breast cancer.
The DNA data put together can help in investigation of disease sources and also reactions of individuals against certain medicines. This is because such baseline data are critical to discovery of genes pertaining to diseases that are of high prevalence in Asia. Genes direct the change in proteins which is the cause of many diseases. About 23% of protein-altering variants found in Asia are unreported in existing databases.
The study published in the journal Nature was conducted by GenomeAsia 100K Consortium, established in 2016 to map the Gap in Genomic Data of which the National Institute of Biomedical Genomics (NIBMG), Kalyani, under the Ministry of Science & Technology is a lead partner.
It is the largest ever genomic study conducted among population groups of Asia. The scientists generated and analysed whole-genome sequences of 1739 individuals from 219 populations spread across most countries of Asia, of whom 598 are from Indian tribal and non-tribal population groups of India.
The team of scientists spanning NIBMG, Genentech, USA, Seoul National University, Korea, Nanyang Technological University, Singapore, recognized that Asian populations were under-represented in global population genomic studies. As a result, appropriate data were unavailable for efficient disease gene discovery in Asian populations and benchmark or baseline data critical to disease gene discovery studies was inadequate.This made DNA microarrays popularly called gene-chips or DNA-chips (an inexpensive technology used for disease gene discovery) relatively difficult.
So far, design of most DNA chips have relied on data primarily of non-Asian individuals. It is known that a point on the DNA that is informative in one population or even in all populations of a geographical region may not be informative in other populations. This is because of variable population ancestries and demographic histories of populations.
The new gene variants identified can make microarray technology more effective for disease investigation. The scientists also say that study can also help identify adverse effects on individuals who possess some specific DNA variants. For example, they found that Carbamezepine – a drug used for treatment of some mental disorders – may have adverse effects on about 400 million speakers of Austronesian languages resident in southeast Asia.
They have found that the structures of populations of Asia are complex and admixed with multiple (fourteen) ancestral populations. However, different regions have admixed with different and smaller subsets of ancestors. They have earlier found that five predominant ancestral admixture events seem to have taken place to give rise to the present-day Indian populations, including the populations of Andaman & Nicobar Islands. Some large urban populations of India like that in Chennai exhibit genetic characteristics that are usually found in isolated populations. They discovered that population mixing occurred on two different occasions with Denisovans in southeast Asia.
Even though Asia comprises about half of the world’s population, less than 10% of genomic data in global databases are from Asian populations. This under-representation has severely constrained efficient conduct of research aimed at identifying genomic bases of diseases of importance in Asian populations.
Further, a large fraction – between 30-40% on an average – of the data generated by the currently available DNA microarrays does not provide useful information on Indian populations. Thus, there is huge amount of wasteful expenditure to assay genotypes of Asians by using these DNA microarrays constructed largely on the basis of data of non-Asians. Deep baseline genomic variation data are required from Asian populations to optimally design DNA microarrays for use in Asian populations. Such data can stimulate the biotechnology industry in Asia. Understanding population structure makes disease gene discovery studies efficient and cost-effective.

Numbers of individuals from various countries of Asia
whose genomes were sequenced and analyzed in this study
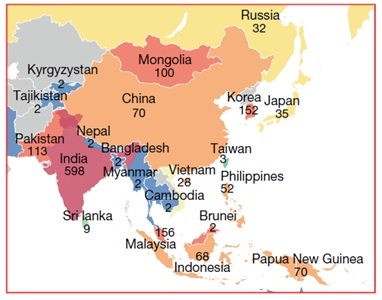
Proposed modern human migration route into southeast Asia during the Last Glacial Maximum about 20,000 years ago with potential locations of admixture of modern humans with the archaic human known as Denisovan (yellow asterisks). Green indicates the above water landmass at the glacial maximum and white outlines indicate present-day shorelines.
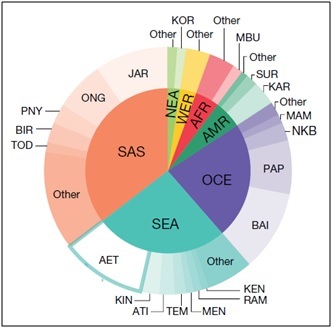
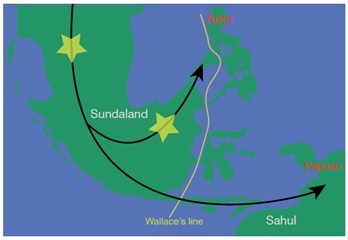
Novel mutations with potential of causing disease identified in populations of various regions of Asia. Such novel mutations were identified in high proportions among tribals of India (Jarawa, JAR; Onge, ONG; Paniya, PNY; Birhor, BIR; Toda; TOD). This indicates that tribal populations may hold the key to identifying DNA changes underlying many genetic diseases