Small molecules called osmolytes help proteins maintain their structure and function under stressful conditions, shows a recent study which provides important insights that could aid in the development of treatments for diseases like Alzheimer's and Parkinson's.
Osmolytes are small molecules that help cells survive stress by stabilizing proteins and preventing them from misfolding. Misfolded proteins can’t perform their functions properly, leading to diseases. Osmolytes are crucial in maintaining the stability of protein structures, making them potential targets for new drugs.
A research team led by Dr. Shubhasis Haldar and his student Deep Chaudhuri at the S.N. Bose National Centre for Basic Sciences- an autonomous institute of Department of Science and Technology used a technique called covalent magnetic tweezers to observe how individual protein molecules fold and unfold under different conditions and interact with osmolytes.
They focused on a protein called Protein L and tested its interaction with two osmolytes-- Trimethylamine N-oxide (TMAO) and trehalose. At higher concentrations, TMAO significantly increased the strength of Protein L, making it more resistant to unfolding.
At low concentrations (up to 1M), TMAO had little effect on the unfolding force of the protein. However, at higher concentrations (1.5M), the unfolding force increased drastically, indicating that TMAO interacts with the folded state of Protein L. This indicates TMAO interacts with the protein in a way that helps it stay folded and stable. High levels of TMAO are linked to heart diseases, so knowing how it interacts with proteins can lead to better treatments.
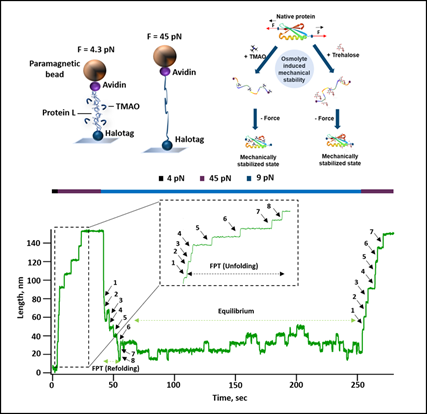
Trehalose, on the other hand, stabilized the unfolded state of the protein, showing that different osmolytes can have varied effects on proteins.
The research published in Nanoscale, a journal of the Royal Society of Chemistry, which can shape understanding of how osmolytes stabilize proteins, will help design better drugs for neurodegenerative diseases and other conditions related to protein misfolding.
Publication link: https://doi.org/10.1039/D3NR00398A